library(rstac)
library(gdalcubes)
library(stars)
library(tmap)
library(dplyr)2: Adding Polygons
We read in a spatial vector dataset with polygons using the Virtual Filesystem Interface (VSI) feature:
url <- "/vsicurl/https://dsl.richmond.edu/panorama/redlining/static/mappinginequality.json"
redlines <-
url |>
st_read() |>
st_make_valid() |>
filter(st_is_valid(geometry))Reading layer `mappinginequality' from data source
`/vsicurl/https://dsl.richmond.edu/panorama/redlining/static/mappinginequality.json'
using driver `GeoJSON'
Simple feature collection with 10154 features and 11 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: -122.7675 ymin: 25.70537 xmax: -69.60044 ymax: 48.2473
Geodetic CRS: WGS 84In sf package, vector data looks almost like a normal tibble (data.frame), but with a special column (usually called “geom” or “geometry”). We can do the usual dplyr operations on the columns, e.g. to select just the redlining polygons in SF:
sf_redlines <- redlines |> filter(city == "San Francisco")We can use this as our bounding box from example 1:
box <- st_bbox(sf_redlines)
start_date <- "2022-06-01"
end_date <- "2022-08-01"
items <-
stac("https://earth-search.aws.element84.com/v0/") |>
stac_search(collections = "sentinel-s2-l2a-cogs",
bbox = c(box),
datetime = paste(start_date, end_date, sep="/"),
limit = 100) |>
ext_query("eo:cloud_cover" < 20) |>
post_request()col <- stac_image_collection(items$features, asset_names = c("B08", "B04", "SCL"))
cube <- cube_view(srs ="EPSG:4326",
extent = list(t0 = start_date, t1 = end_date,
left = box[1], right = box[3],
top = box[4], bottom = box[2]),
dx = 0.0001, dy = 0.0001, dt = "P1D",
aggregation = "median", resampling = "average")
mask <- image_mask("SCL", values=c(3, 8, 9)) # mask clouds and cloud shadows
data <- raster_cube(col, cube, mask = mask)ndvi <- data |>
select_bands(c("B04", "B08")) |>
apply_pixel("(B08-B04)/(B08+B04)", "NDVI") |>
reduce_time(c("mean(NDVI)")) ndvi_stars <- st_as_stars(ndvi)mako <- tm_scale_continuous(values = viridisLite::mako(30))
fill <- tm_scale_continuous(values = "Greens")
tm_shape(ndvi_stars) + tm_raster(col.scale = mako) +
tm_shape(sf_redlines) + tm_borders("grade", lwd=2)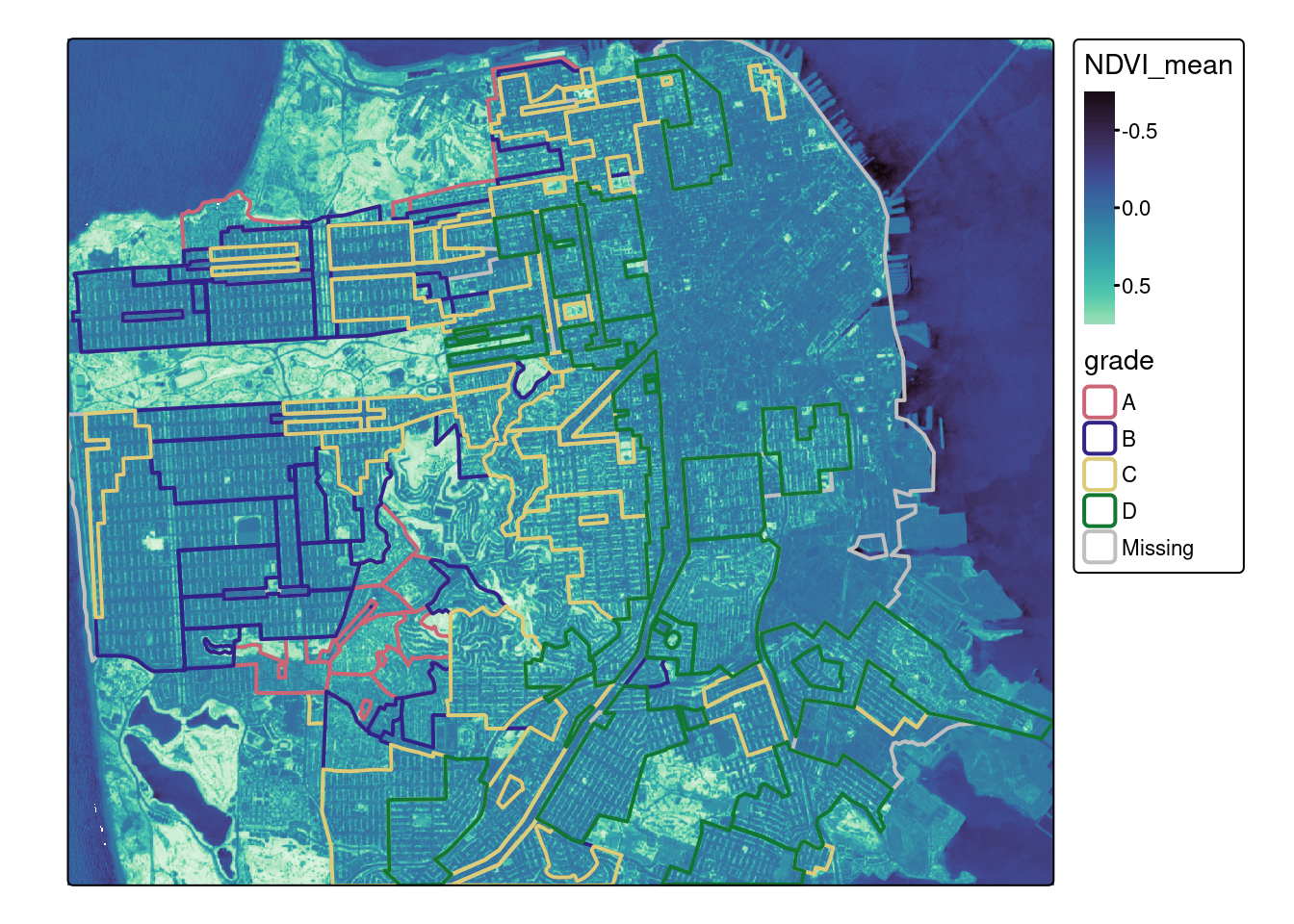
Interactive maps
tmap_mode("view")tmap mode set to 'view'tm_shape(ndvi_stars) + tm_raster(col.scale = mako) +
tm_shape(sf_redlines) + tm_borders("grade", lwd=2)